UNM research at SC17
The Center for Advanced Research Computing will take a collection of digital posters highlighting computational research at UNM and partner New Mexico State University to SC17 in Denver in November. Topics range from race car aerodynamics to cancer nanotherapy. Posters will be displayed on TV monitors at the UNM CARC booth #535 on the SC17 exhibition floor.
UNM research posters
CARC helps UNM race car team’s need for speed
 The University of New Mexico Formula SAE is making race cars go faster with the help of the university’s Center for Advanced Research Computing (CARC). Formula SAE is an international engineering-design competition where student teams from universities design and build a Formula-1/Indy style race car to compete against other universities. The cars are judged on many aspects including design, cost, acceleration, autocross, and endurance. In the three-semester program at UNM, student teams design, build, and race a car in international competition.
The University of New Mexico Formula SAE is making race cars go faster with the help of the university’s Center for Advanced Research Computing (CARC). Formula SAE is an international engineering-design competition where student teams from universities design and build a Formula-1/Indy style race car to compete against other universities. The cars are judged on many aspects including design, cost, acceleration, autocross, and endurance. In the three-semester program at UNM, student teams design, build, and race a car in international competition.
Determination and characterization of clinically obtained viral strains via next generation sequencing and post sequencing bioinformatic analysis
The research focus of Dr. Darrell Dinwiddie's lab is to use genomic medicine (a structured approach to disease diagnosis and management that prominently features next-generation sequencing and analysis at a genome scale) to diagnose, discover, and investigate rare monogenic genetic disorders and to apply genomics to understand immune-mediated disorders.
Gastropod comparative genomics
The biology and evolution of phylogenetic groups of animals is often interpreted through the proxy of a few well-studied model organisms. Next generation sequencing yields broad access to genomes and facilitates novel gene discovery and evaluation of the predictive value of model species for larger animal clades.
Gene expression signatures of acute respiratory syncytial virus infection in pediatric patients reveals insight into clinical pathogenesis
The research focus of Dr. Darrell Dinwiddie's lab is to use genomic medicine (a structured approach to disease diagnosis and management that prominently features next-generation sequencing and analysis at a genome scale) to diagnose, discover, and investigate rare monogenic genetic disorders and to apply genomics to understand immune-mediated disorders.
Genomic comparisons of multipartite symbiosis: Understanding the metabolic basis of parasitism
Massive amounts of sequence data are being generated daily, coming from metagenome projects and microbiome projects. A major challenge today is how to make sense of these data. We are mainly concerned with interactions among organisms existing in multipartite symbioses. By trying to answer questions regarding symbioses (encompassing both mutualistic and parasitic) through understanding symbiosis as an emergent property of the metabolic network/metabolic capacity of the whole system. Marked by genome reduction and gene loss, most parasites lose metabolic functions as they increase their dependence on nutrients offered by the host. The question is then: do parasites from different phyla converge in their lost/gained metabolic functions as they adapt to a parasitic life style? By understanding the evolutionary patterns behind the metabolic networks of these parasites we can evoke new methods to combat some of these medically important species.
keybin: Key-based binning for distributed clustering
keybin is a distributed key-based binning clustering algorithm for high-dimensional spaces. keybin locally generates a spatial key for each data point across all dimensions without needing knowledge of other data. Then, it performs a conceptual Map-Reduce procedure in the index space to learn a global clustering structure with limited communication. It can scale with the dimensionality and size of data sets and preserve privacy at the same time.
Mountain lions on the edge: Integrating conservation into urban planning through predictive modeling
 Can city dwellers and wildlife coexist safely and peacefully? UNM School of Architecture and Planning graduate student Sophia Thompson is convinced they can. She is using the UNM Center for Advanced Research Computing resources to do research on New Mexico mountain lions, with the goal of helping humans and wildlife coexist.
Can city dwellers and wildlife coexist safely and peacefully? UNM School of Architecture and Planning graduate student Sophia Thompson is convinced they can. She is using the UNM Center for Advanced Research Computing resources to do research on New Mexico mountain lions, with the goal of helping humans and wildlife coexist.
Onload vs offload for many-core architectures
In this study, researchers examined the power and performance tradeoff of Network offload. By utilizing a cluster outfitted with custom power monitoring hardware and isolating the hardware changes to the network card they were able to precisely measure power and performance differences between the a Mellanox offload card and a Qlogic onload card. Utilizing dynamic voltage and frequency scaling, they were able to approximate the performance to evaluate potential impacts on a wide array of processors.
Microbenchmark results show an increase in performance when using offload cards as the CPU is not a bottleneck in these cases. Offload takes less power at high frequencies and more power at lower frequencies. Examining component level power measurements, the offload network card leads to an increase power use, while the CPU uses less due to decreased activity.
Optical rogue wave generation in dielectrics with correlated fluctuations of refractive index
Rogue waves (RWs) are extreme high intensity waves that seem to appear randomly. RWs were first studied in the context of oceanography to explain the presence of huge and destructive waves in relatively calm water. These studies have since been extended to other physical systems like microwaves, nonlinear crystals, cold atoms, Bose-Einstein condensates, and even in non-physical systems like the financial market. There is still an ongoing debate on whether the RWs are originated due to linear or nonlinear processes. In this work, researchers explore RW generation for the propagation of light in a dielectric in the presence of correlated fluctuations of its refractive index. They observe that with increasing the correlation length, the probability of RW generation increases. This result is important because in practice most fluctuations occur with a finite correlation length and it appears that it is important to take this into account to get a realistic account of RW generation.
RAMP: The Repository Analytics and Metrics Portal
Institutional repositories (IR) contribute to the scholarly ecosystem by providing efficient, open access to the research products of a university or organization. They further provide a means for researchers and administrators to assess research impact via metrics including file download counts. Unfortunately, a lack of consistency among repository and analytics platforms casts doubt upon the reliability of such statistics. RAMP is an easily configured web application which integrates with the Google Search Console API to allow participating IR administrators access to consistent, reliable statistics about repository usage. Because the same data and analytics models are applied across participating repositories, the RAMP prototype facilitates the establishment of baseline statistics for comparisons between similar repositories.
RMA-MT: A benchmark suite for assessing Message Passing Interface (MPI) multi-threaded RMA performance
RMA-MT is a novel benchmark suite that examines the combination of two proposed exascale parallel programing paradigms. Multi-threading (MT) has been suggested in the MPI+X models to deal with the high levels of intra-node parallelism, particularly when dealing with many-core processors. Remote Memory Access (RMA) has been proposed as a communication interface to to provide shared memory abstractions which bypass expensive point-to-point data structures.
Researchers seek ‘magic bullet’
 UNM researchers aim to deliver chemotherapy directly to cancerous tissue, working to encapsulate toxic chemotherapy in nanoparticles. The challenge is that the body’s immune system interprets nano-drug-carriers as foreign bodies, attacking and diverting them. Researchers are using CARC to search for a solution.
UNM researchers aim to deliver chemotherapy directly to cancerous tissue, working to encapsulate toxic chemotherapy in nanoparticles. The challenge is that the body’s immune system interprets nano-drug-carriers as foreign bodies, attacking and diverting them. Researchers are using CARC to search for a solution.
Smart molecular devices for sensing and diagnostics
How can scientists design nanoscale sensors and devices to diagnose and treat disease? Dr. Matthew Lakin, UNM Assistant Professor of Computer Science, is using UNM’s computational resources to design synthetic DNA computing devices, which can detect molecular inputs such as disease biomarkers, and make diagnostic decisions based on them. This work is in collaboration with the UNM Center for Biomedical Engineering.
Space exploration: Taken by Swarm
If humans one day colonize Mars, perhaps the University of New Mexico will have played a small part. UNM students were among those from across the country participate in the NASA Swarmathon robotics competition. Learn more about this event from UNM computer science professor Melanie Moses.
Uncertainty quantification for seismic events: Extracting uncertainty from seismic waveforms through parametric bootstrapping
Can scientists do a better job at detecting small seismic events? UNM Department of Computer Science Ph.D. student Matt Peterson thinks there is a way. Working at Sandia National Labs, he is using a combination of Physics and Statistics to research a way to produce a probability distribution for a Seismic Signal Arrival time.NMSU research posters
A phylogenetically informed investigation of transcriptome evolution and transcriptome response of the tropical crop genus Leucaena to psyllid herbivores
 The extensive application of next generation sequencing in agricultural research has focused on yield improvements in the most important global food crops. However, lesser-known crops receive comparatively little research attention, such as the genus Leucaena. Leucaena can provide protein-rich leaves for livestock feed; green manure used as nitrogen-rich fertilizer for crops and soil stabilization, and rapidly renewable woody biomass for construction, fuelwood, and even biofuels. One of the goals of this project is to investigate how plant transcriptomes change through time and in association with diversification of species.
The extensive application of next generation sequencing in agricultural research has focused on yield improvements in the most important global food crops. However, lesser-known crops receive comparatively little research attention, such as the genus Leucaena. Leucaena can provide protein-rich leaves for livestock feed; green manure used as nitrogen-rich fertilizer for crops and soil stabilization, and rapidly renewable woody biomass for construction, fuelwood, and even biofuels. One of the goals of this project is to investigate how plant transcriptomes change through time and in association with diversification of species.
Automated image analysis for segmenting and classifying New Mexico desert plants
The scope of the entire project is to determine if Criollo cattle are beneficial to the land at the Jornada Experimental Range. A camera found on the cow’s collar captures images and tracks the vegetation that the Criollo cattle are consuming. A mixture of image processing and machine learning are used to correctly classify and identify the plant species found in the images.
BIGDATA: Collaborative research: F: Discovering context-sensitive impact in complex systems
Successfully tackling many urgent challenges in socio-economically critical domains (such as sustainability, public health, and biology) requires obtaining a deeper understanding of complex relationships and interactions among a diverse spectrum of entities and agents in different contexts. While some of the relationships (e.g., co-location of energy production facilities and water delivery networks in the energy domain and scheduled flights between two cities in the study of epidemics) in these domains are explicitly known, the knowledge of these explicit relationships is often far from sufficient to enable decision making. What is required instead is an understanding of whether and (if so) in what contexts these entities impact each other. The goal of this project is to establish the theoretical, algorithmic, and computational foundations of big data-driven Context-Sensitive Impact Discovery (CSID) in complex systems.
Data Storage Lab @NMSU
Dr. Mai Zheng's research interests are in the broad areas of computer systems. He started the Data Storage Lab@NMSU to play with the latest storage technologies and strive to advance the reliability and performance for data, for people.
New computational chemistry methods to quantify the properties of atoms in materials
Atomistic properties help us understand material properties and chemical reactivity. Netatomic charges quantify electron charge transfer between atoms. Atomic spin moments quantify the magnetic properties of materials at the atomistic level. Bond orders quantify the number of electrons exchanged between two atoms in a material. Polarizability quantifies the electrical response of a material to an applied electric field. Dispersion coefficients describe the van der Waals forces that hold materials like non-polar liquidstogether. This project develops more accurate methods to compute these atomistic descriptors using the quantum mechanically computed electron and spin density distributions of the material as input. The methods are computationally efficient and canbe applied to a wide variety of material types.
The Hilbert Spectrum: A general framework for time-frequency analysis
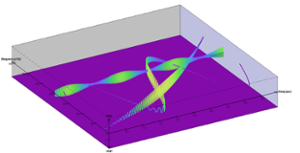 This research is a contribution to the old problem of representing a signal in the coordinates of time and frequency. We define a signal model composed of a superposition of complex, AM-FM components that enables rigorous definition of instantaneous amplitude and instantaneous frequency. Based on this model, the Hilbert Spectrum (HS) was introduced as the ideal instantaneous spectrum, which specializes to the Fourier spectrum. Numerical algorithms are given for computing a HS where the component is assumed to be a latent intrinsic mode function. Researchers demonstrate that the HS simultaneously exhibits fine resolution in both the coordinates of time and frequency simultaneously.
This research is a contribution to the old problem of representing a signal in the coordinates of time and frequency. We define a signal model composed of a superposition of complex, AM-FM components that enables rigorous definition of instantaneous amplitude and instantaneous frequency. Based on this model, the Hilbert Spectrum (HS) was introduced as the ideal instantaneous spectrum, which specializes to the Fourier spectrum. Numerical algorithms are given for computing a HS where the component is assumed to be a latent intrinsic mode function. Researchers demonstrate that the HS simultaneously exhibits fine resolution in both the coordinates of time and frequency simultaneously.
REU site: BIGData: Big data analytics for cyber-physical systems
The goal of this Research Experience for Undergraduates (REU) site initiative (BIGDatA-BIG Data Analytics for Cyber-physical Systems) is to inspire and prepare undergraduate students to pursue academic studies and careers in STEM with a focus on big data analytics for cyber-physical systems (CPS). The site’s research projects will focus on equipping students with skills and knowledge related to big data analytics in CPS. The CPS will be exemplified by four main application areas: smart grids, wireless sensor networks, smart homes for the elderly and disabled, and disaster response.
The technicolor dawn simulations
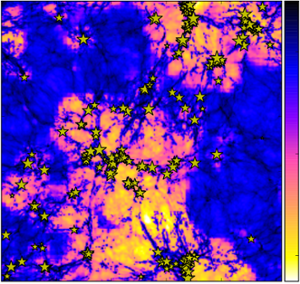 The way in which gas, galaxies, and light interacted during the first 1.5 billion years is a frontier both for theoretical models and for observations. How did those faint, early galaxies go about forging gas into stars, irradiating the Universe with energetic light, and enriching their environments with the heavy elements that dominate our lives? This project, led by Kristian Finlator, Assistant Professor of Astronomy at NMSU, combines updated models for the formation of stars and galaxies with a multifrequency radiation transport solver to compute simultaneously the formation of young galaxies, their environments, and the growth of the nascent extraviolet ultraviolet background.
The way in which gas, galaxies, and light interacted during the first 1.5 billion years is a frontier both for theoretical models and for observations. How did those faint, early galaxies go about forging gas into stars, irradiating the Universe with energetic light, and enriching their environments with the heavy elements that dominate our lives? This project, led by Kristian Finlator, Assistant Professor of Astronomy at NMSU, combines updated models for the formation of stars and galaxies with a multifrequency radiation transport solver to compute simultaneously the formation of young galaxies, their environments, and the growth of the nascent extraviolet ultraviolet background.
