Researchers use computer modeling to understand COVID-19 infection mechanism
New Mexico Consortium Researcher Wataru Nishima, in collaboration with University of Warsaw Assistant Professor Marta Kulik, has authored a paper detailing new insights into the infection mechanism of COVID-19. The team used Center for Advanced Research Computing systems to develop a novel computational model of SARS-CoV-2, the virus that causes COVID-19, and simulate the way it attaches to host cells. Nishima and Kulik have proposed a new hypothesis regarding the process of SARS-CoV-2 cell infection, which could lead to more effective treatment and prevention of COVID-19.
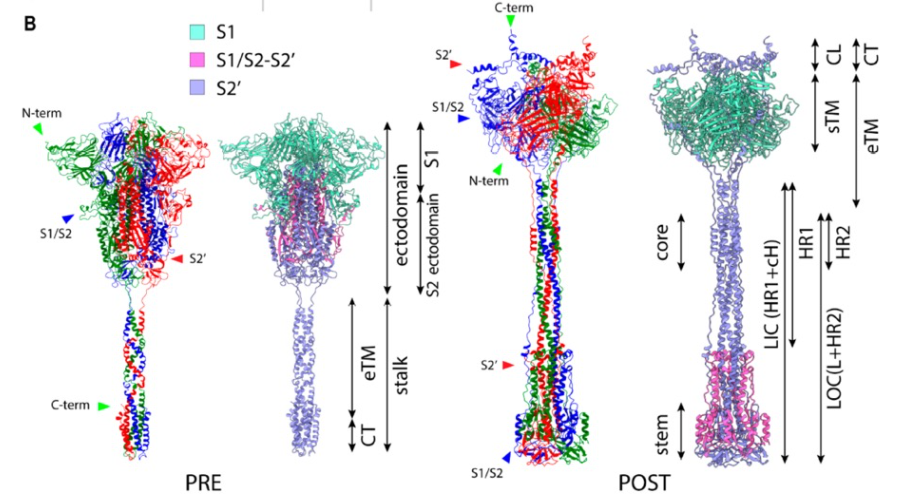
While scientists know that the SARS-CoV-2 virus infects cells by using a spike protein (a barb-shaped structure located on the outside of the virus) to fuse its own membrane with that of the host cell, the details of this process are not yet fully understood. Experimental studies have revealed much of the structure of the virus’ spike protein before and after fusing with a host cell membrane, leading to a widely accepted model of the membrane fusion process. This model, however, is not conclusive and cannot explain some observed characteristics of the virus.
To reconcile this inconsistency, Nishima and Kulik created computer simulations to model the way a SARS-CoV-2 spike protein invades a host cell. The surprising results of the team’s simulations suggest a different mechanism of SARS-CoV-2 infection than had been previously hypothesized. According to the widely accepted model of SARS-CoV-2 cell invasion, the spike protein should remain outside of the host cell’s membrane, while Nishima and Kulik’s simulations show that the spike protein may actually enter cells. The team’s new model of infection is consistent with experimental data and explains some of the observed phenomena that cannot be explained by the widely accepted model—namely, the way antibodies prevent SARS-CoV-2 entry and the differing characteristics of infectivity in some strains of the virus.
If true, this hypothesis could substantially change the way we approach the coronavirus pandemic. Nishima is careful to note, however, that much more research is needed to investigate these findings, writing, “This is a computational work… proposing a hypothesis about the initial infection mechanism that is implicated by the computational structural model and experimental knowledge. To get conclusive ideas, it has to be validated by experiments.”
The team’s computer simulations were performed using CARC resources. Nishima explains that using computational modeling has advantages over wet lab experimentation in some cases; computational modeling is typically cheaper and safer, can be performed remotely, and allows researchers to execute a greater number of trials using automated workflows.
The authors of this study hope that their simulations will lead to further research and, ultimately, improved treatment and prevention of coronavirus infection. Nishima urges other scientists to get involved in COVID-19 research, writing, “I would like to encourage researchers to participate [in] the fight against COVID with their expertise. To tackle [this] highly complex problem, integration/collaboration of scientific efforts from different area[s is] now highly desired.”
Image: A representation of Nishima and Kulik’s computational model of the SARS-CoV-2 spike protein before and after fusion with a host cell membrane, courtesy of Wataru Nishima.
